Discretizing the disk
3.3. Discretizing the disk#
The unit disk is a tensor-product domain—with a twist. Usually we think of \(0\le r \le 1\) and \(-\pi\le \theta < \pi\), but that formulation leads to an artificial singularity at the origin which then has to be remedied. It’s better to start with
Choose discretization sizes and let
In order to avoid trouble at \(r=0\), we require that \(n_r\) is odd.
If \(\bfU\) is a matrix of unknowns on a grid in \((r,\theta)\) space, then differentiation in \(r\) is straightforward: \(\bfD_r \bfU\). With respect to \(\theta\), however, there is an implicit periodicity we have to respect: the row corresponding to \(r=-1+kh\) should continue smoothly into the row at \(r=1-kh\), and vice versa, to form data around one actual circle inside the unit disk. Let
Then
double-covers the disk but has rows with true periodicity. Thus, \(\bfD_{\theta}\), \(\bfD_{\theta\theta}\), and so on can be defined in the usual way for periodic conditions on a discretization size \(2n_{\theta}\), e.g.,
After differentiation, though, we want to discard the duplicated information and return to the standard discretization, which means discarding the last \(n_\theta\) columns. Let
be a partitioning into \(n\times n\) blocks. Then the full differentiation process can be expressed as
Following this process term by term for a linear differential operator leads to a Sylvester-style equation that we can transform using Kronecker products.
Example 3.3.1 (Polar Laplacian)
The Laplacian in polar coordinates is
Therefore, a discretization of this operator on the unit disk is
where \(\bfS\) is a diagonal matrix of the values \(1/r_i\) over all \(n_r\) nodes. From here, things proceed as they did before. For a Poisson problem, this can be equated to a given grid function \(\bfF\), at least before imposing boundary conditions (only at \(r=\pm 1\)). In order to use direct linear algebra, we can transform the equation to standard form using Kronecker products; for a Krylov subspace iteration, we can work directly with the form above.
include("diffmats.jl")
function polarlap(nr,nθ)
@assert isodd(nr)
⊗ = kron
r,Dr,Drr = diffmats(nr,-1,1)
S = spdiagm(1 ./r)
q = π/nθ
θ = q*(0:nθ-1)
Dθθ = 1/q^2*spdiagm(
0=>fill(-2.,2nθ),
-1=>ones(2nθ),
1=>ones(2nθ),
2nθ-1=>[1.],
1-2nθ=>[1.]
)
Q₁₁,Q₁₂ = Dθθ[1:nθ,1:nθ],Dθθ[nθ+1:2nθ,1:nθ]
return r,θ,I(nθ)⊗(Drr + S*Dr) + Q₁₁⊗S.^2 + Q₁₂⊗reverse(S.^2,dims=2)
end
using Plots
r,θ,L = polarlap(11,20)
spy(L)
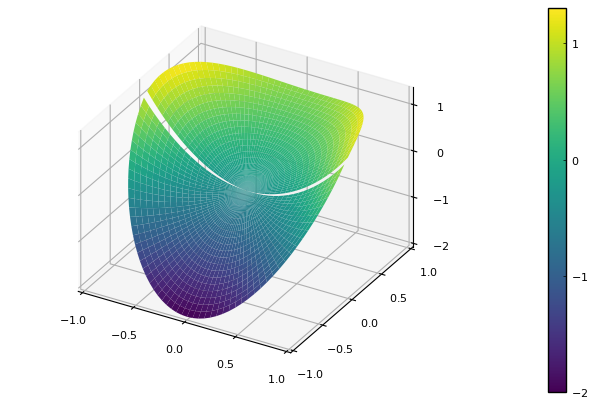
û(x,y) = x.^2 + 2y
f(x,y) = 2
nr,nθ = 39,58
r,θ,A = polarlap(nr,nθ)
X = [r*cos(θ) for r in r, θ in θ]
Y = [r*sin(θ) for r in r, θ in θ]
bdy = falses(nr+1,nθ)
bdy[1,:] .= true
bdy = vec(bdy)
u = zeros(size(A,1))
u[bdy] = û.(X[bdy],Y[bdy])
inter = @. !bdy
à = A[inter,inter]
f̃ = f.(X[inter],Y[inter]) - A[inter,bdy]*u[bdy]
u[inter] = Ã\f̃;
pyplot()
U = reshape(u,nr+1,nθ)
surface(X,Y,U,color=:viridis)